Common wisdom states that a new pathogen, once introduced into a vulnerable human population with no immune defenses, will evolve over time to grow more benign and live in amity with its host. After all, the argument goes, a pathogen will depend on its host for its own survival, so causing mass disease and death will make onward transmission more difficult and ultimately only ensure its own demise.
Unfortunately, despite the reassuring logic, the reality is often more nuanced. Infection by water or food-borne pathogens can commonly result in severe or even life-threatening diarrhea. However, far from disadvantaging the pathogen, this symptom can actively facilitate onward transmission via the fecal-oral route. Malaria is a vectored disease (those that are transmitted by living organisms) that is spread via mosquitos. In this case, the extreme sickness suffered by the host won’t impede onward transmission if there are plenty of healthy mosquitos around to help the parasite jump ship from one host to the next.
So how might we expect the novel coronavirus SARS-CoV-2 to evolve as it adjusts to its new way of life, infecting and spreading among humans? Will the symptoms naturally ameliorate while the pandemic eventually fizzles out? Or are the fears of more virulent strains emerging and sparking a deadly second wave justified? In the interests of expectation management, the short answer is that we don’t know.
In a seminal work from 1991, Roy Anderson and Robert May argued that it is near-impossible to predict the future trajectories of virulence and transmissibility of emerging pathogens. However, although soothsaying remains out of our reach, modern technology has provided one critical edge for the real-time monitoring of natural mutations in viruses. This critical edge is genome sequences — and lots of them.
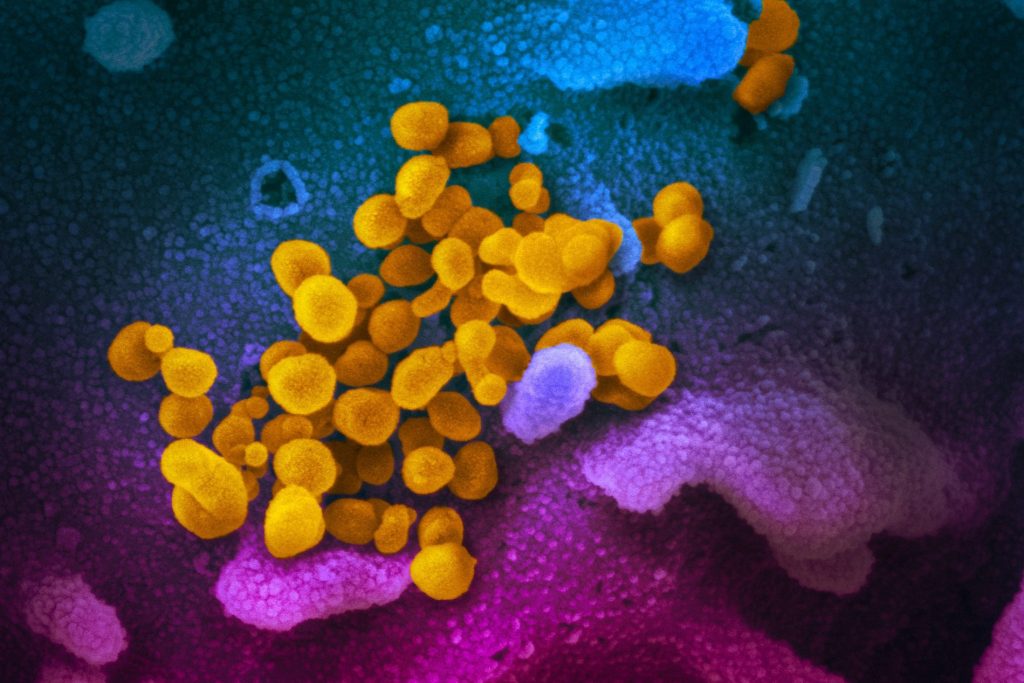
Whether you consider them living or not, viruses — like all biological entities — are in essence a string of genetic instructions encoded by a genome made of DNA (or, in this case, RNA which is a close chemical cousin). The SARS-CoV-2 genome is under 30,000 bases (or letters) long and encodes only 15 genes, making it only 1 percent the size of a typical bacterial genome and 0.0001 percent the size of a human genome.
Such a small genetic package can be easily sequenced and over 45,000 SARS-CoV-2 genomes are now available, over half of which were generated in the UK. These data tell us that the virus evolved in bats and entered the human population only once, possibly via an intermediate animal, between late October and early December last year. We know that the viral genome is mutating, as all genomes do, but at a relatively slow rate — around one mutation every two weeks. This makes SARS-CoV2 more stable than its close relative SARS, and much more stable than the viruses that cause flu.
Mutations arise as the virus replicates and new, slightly imperfect, copies are made of the genetic material. Being random, the consequences for the virus of these mutations can vary — they can be harmful, beneficial or neither. Most mutations will fall into the final category, and these ‘neutral’ mutations will have no material consequence for how the virus behaves. Whether viruses carrying these mutations will go extinct or spread further is down to pure chance.
Many of the early reports of different strains of the virus from across the world were in fact based only on these functionally irrelevant changes that don’t actually do anything. Neutral mutations are, however, of great practical importance. By acting as molecular flags on the genome, they make it possible for epidemiologists to understand how the virus is spreading both globally and locally. For example, by combining genome sequences with travel data, it is known that there were at least 1,356 separate introductions of the virus in the UK during March, mostly from Spain, France and Italy. As the pandemic progresses, such approaches will play a critical role in identifying localized outbreaks, as the patterns of mutations on all the genomes from cases linked to a single source should be identical.
So, if most mutations are neutral, how can we tell which ones, if any, are likely to change the way the virus behaves? There are two key lines of evidence from the genome sequences; one is how often the same mutation is observed and the second is which specific gene has been changed by the mutation and how.
[special_offer]
Consider a random mutation that increases the rate at which the virus will spread, or improves the viruses adaption to the human host in some other way. Almost by definition, we will see more strains carrying this mutation than we would expect by chance. However, more tellingly, we might also expect the same mutation to arise in the virus independently on more than one occasion, in unrelated lineages and in different geographical locations. This does not mean that the mutation is somehow deliberate, it is still a random event, it is simply the case that when such a mutation arises by chance it is much more likely to spread and thus be observed than neutral mutations.
Once such recurrent mutations have been identified, the next step is to consider where they are in the genome and what the biological consequence might be. For example, recurrent mutations have been identified that are predicted to change a critical part of the protein that enables the virus to bind to and invade human cells. The final and most difficult step is to carry out the necessary experiments to prove the significance of the mutation in the laboratory.
The current evidence remains equivocal on whether the virus is mutating to become more or less of a threat, and we have no epidemiological framework for robustly predicting whether it will. Nevertheless, the wealth of genome data, at the very least, provides a means to rapidly identify those mutations that are likely to change the way the virus behaves. From there, we can improve our therapeutic strategies and better manage this disease.
This article was originally published on the University of Bath IPR blog.